Homework 3¶
Imports¶
# import relevant packages
import numpy as np
from scipy.linalg import solve
import matplotlib.pyplot as plt
from matplotlib import rcParams
import serpentTools
from serpentTools.settings import rc
from plotter import Plot1d
from diffusion_coeffs import *
# Default values
FONT_SIZE = 16 # font size for plotting purposes
# rcParams['figure.dpi'] = 300
plt.rcParams['figure.figsize'] = [6, 4] # Set default figure size
Question 1 - basic transport xs calculations¶
# Define file and read file
resFile = "./serpent_HW3/fa2D_70gr_inf_res.m"
rc["serpentVersion"] = "2.1.32"
res = serpentTools.read(resFile)
SERPENT Serpent 2.2.1 found in ./serpent_HW3/fa2D_70gr_inf_res.m, but version 2.1.32 is defined in settings
Attempting to read anyway. Please report strange behaviors/failures to developers.
ng = 70
univ0 = res.getUniv('0', timeDays=0)
flx = univ0.infExp['infFlx']
sigT = univ0.infExp['infTot']
SP1 = univ0.infExp['infSp1']
cmmTransp = univ0.gc['cmmTranspxs'] # represents in-scatter
infTransp = univ0.infExp['infTranspxs'] # out-scatter
energy = univ0.groups * 1E+06
SP1=SP1.reshape((ng,ng)).transpose()
# Inscattering
transportxs, tau, Jg = InScatter(ng=70, sigS1=SP1, sigT=sigT, flx=flx, B2=0.0001) # get inscatter
D = Condense2gr(xs=1/3/transportxs, flx=flx,energy=energy)
print("Inscatter Diffusion Coeff. =", D)
# Outscatter
transportxs, tau = OutScatter(ng=70, sigS1=SP1, sigT=sigT)
D = Condense2gr(xs=1/3/transportxs, flx=flx,energy=energy)
print("Outscatter Diffusion Coeff. =", D)
# Flx Limited
transportxs, tau = FluxLimited(ng=70, sigS1=SP1, sigT=sigT, flx=flx)
D = Condense2gr(xs=1/3/transportxs, flx=flx,energy=energy)
print("Flux Limited Diffusion Coeff. =", D)
Inscatter Diffusion Coeff. = [1.69854949 0.88303605]
Outscatter Diffusion Coeff. = [1.73376106 0.82629467]
Flux Limited Diffusion Coeff. = [1.68208348 0.87193724]
Question 2¶
Step 1. Get \(\Sigma_{tr,g}^{H,in}\) and \(\tau_{H,in}\)¶
# Define file and read file
resFile = "./serpent_HW3/infinite_h_basic_h1_r1_res.m"
rc["serpentVersion"] = "2.1.32"
res = serpentTools.read(resFile)
ng = 70
univ0 = res.getUniv('0', timeDays=0)
flx = univ0.infExp['infFlx']
sigT_H = univ0.infExp['infTot']
SP1 = univ0.infExp['infSp1']
cmmTransp = univ0.gc['cmmTranspxs'] # represents in-scatter
infTransp = univ0.infExp['infTranspxs'] # out-scatter
energy = univ0.groups * 1E+06
SP1=SP1.reshape((ng,ng)).transpose()
# In scattering xs
transportxs_H_in, tau_in, _ = InScatter(ng=70, sigS1=SP1, sigT=sigT_H, flx=flx)
transportxs_H_out, tau_out= OutScatter(ng=70, sigS1=SP1, sigT=sigT_H)
Now get transport xs for the FA¶
# Now get inscattering and outscattering for fuel assembly
res_assy = serpentTools.read('serpent_HW3/fa2D_70gr_inf_res.m')
rc["serpentVersion"] = "2.1.32"
ng = 70
univ0 = res_assy.getUniv('0', timeDays=0)
flx = univ0.infExp['infFlx']
sigT = univ0.infExp['infTot']
SP1 = univ0.infExp['infSp1']
cmmTransp_fuel = univ0.gc['cmmTranspxs'] # represents in-scatter
cmm_tau_fuel = cmmTransp_fuel / sigT
energy = univ0.groups * 1E+06
SP1=SP1.reshape((ng,ng)).transpose()
transportxs_fuel_out, tau_fuel_out = OutScatter(ng=70, sigS1=SP1, sigT=sigT)
transportxs_fuel_in, tau_fuel_in, _ = InScatter(ng=70, sigS1=SP1, sigT=sigT, flx=flx, B2=1e-3)
SERPENT Serpent 2.2.1 found in serpent_HW3/fa2D_70gr_inf_res.m, but version 2.1.32 is defined in settings
Attempting to read anyway. Please report strange behaviors/failures to developers.
# get area of water here.
area_tot = 21.5**2
area_fuel = 264 * (np.pi * 0.475**2)
area_guide = 25 * np.pi * (0.6120**2 - 0.5715**2)
area_water = area_tot - area_fuel - area_guide
# Number density of water in water only simulation
Na = 6.02214076e23
Nh_inf = Na * 0.7129 / 1.00782503223
N_water = (Na * 0.7 / (1.00782503223*1 + 2*15.999) ) # water number density
Nh_lattice = 1 * N_water* area_water/area_tot # Number density of H in lattice
# Out-scattering transport xs from hydrogen only simulation -> transportxs_H_out
# Out-scattering transport xs from fuel assembly simulation -> transportxs_fuel_out
# Transport correction factor from Inscattering on hydrogen -> tau_in
# Step 1. get micro-xs from inf hydrogen simulation (bullet 2 or step 1)
micro_transport_H_out = transportxs_H_out / Nh_inf
# Step 3. equation 7.14
transport_lattice_without_H = transportxs_fuel_out - micro_transport_H_out*Nh_lattice
# Step 4. Use in-scattering tau to correct total xs for hydrogen to get transport corrected hydrogen
transport_hydrogen_only_corrected = sigT_H / Nh_inf * Nh_lattice * tau_in
# Step 5. Hydrogen correction Equation 7.16
transport_corrected_via_hydrogen = transport_lattice_without_H + transport_hydrogen_only_corrected
# Final - get diffusion coeff. by collapsing
hydrogen_corrected_diff_2g = Condense2gr(1.0/3.0/transport_corrected_via_hydrogen, flx, energy, cutoffE=0.625)
print("Hydrogen correction diffusion coeff. =", hydrogen_corrected_diff_2g)
Hydrogen correction diffusion coeff. = [1.72253227 0.8400543 ]
plt.figure(figsize=[6,4])
# Plot1d(energy, tau_in, xlabel="Energy, eV", ylabel='',
# fontsize=16, marker="-k", markerfill=False, markersize=6)
# Plot1d(energy, tau_out, xlabel="Energy, eV", ylabel='',
# fontsize=16, marker="-r", markerfill=False, markersize=6)
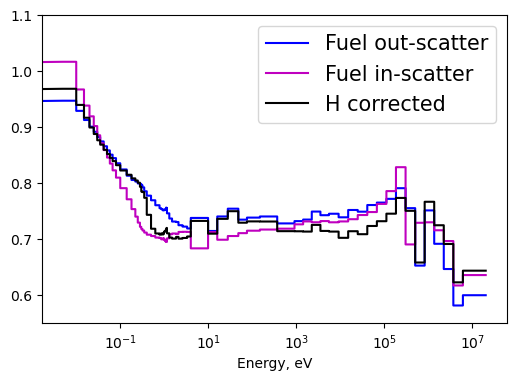
Plot1d(energy, tau_fuel_out, xlabel="Energy, eV", ylabel='',
fontsize=16, marker="-b", markerfill=False, markersize=6)
Plot1d(energy, tau_fuel_in, xlabel="Energy, eV", ylabel='',
fontsize=16, marker="-m", markerfill=False, markersize=6)
# Plot1d(energy, cmm_tau_fuel, xlabel="Energy, eV", ylabel='',
# fontsize=16, marker="r-", markerfill=False, markersize=6)
Plot1d(energy, transport_corrected_via_hydrogen/sigT, xlabel="Energy, eV", ylabel='',
fontsize=16, marker="k-", markerfill=False, markersize=6)
plt.legend(['Fuel out-scatter', 'Fuel in-scatter','H corrected'], fontsize=15)
plt.ylim([0.55,1.1])
plt.grid()
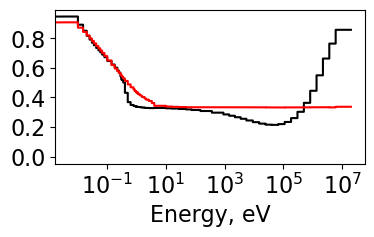
# Plot tau from inscattering and outscattering hydrogen corrections
plt.figure(figsize=[4,2])
Plot1d(energy, tau_in, xlabel="Energy, eV", ylabel='',
fontsize=8, marker="-k", markerfill=False, markersize=6)
Plot1d(energy, tau_out, xlabel="Energy, eV", ylabel='',
fontsize=8, marker="-r", markerfill=False, markersize=6)
Question 5: Getting CM transport xs and diffusion coefficients with CM method implementation¶
# Define file and read file
resFile = "./serpent_HW3/fa2D_70gr_inf_res.m"
rc["serpentVersion"] = "2.1.32"
res = serpentTools.read(resFile)
ng = 70 # number of energy groups
univ0 = res.getUniv('0', timeDays=0)
sigT = univ0.infExp['infTot']
cmmTransp = univ0.gc['cmmTranspxs'] # represents in-scatter (cumulative migration method)
infTransp = univ0.infExp['infTranspxs'] # out-scatter ()
infinite_flx = univ0.infExp['infFlx']
infinite_flx = infinite_flx/infinite_flx.sum()
rabsxs = univ0.infExp['infRabsxs']
nusigF = univ0.infExp['infNsf']
chi = univ0.infExp['infChit']
SP0 = univ0.infExp['infSp0']
SP1 = univ0.infExp['infSp1']
kinf = res.resdata['absKeff'][0] # k-in
energy = univ0.groups * 1E+06
SP0=SP0.reshape((ng,ng)).transpose()
SP1=SP1.reshape((ng,ng)).transpose()
# Get the CM flux and b2
cm_flx, b2_cm = CMSpectrum(ng=ng, sigMtx0=SP0, sigT=sigT, sigTr=cmmTransp, chi=chi, nuSigF=nusigF, kinf=kinf)
b1_flux, b1_ij, b2_b1 = CriticalSpectrum(ng, SP0, SP1, sigT, chi, nusigF, kinf)
p1_flux, p1_ij, b2_p1 = CriticalSpectrum(ng, SP0, SP1, sigT, chi, nusigF, kinf, P1=True)
print()
print("CM B2 =", b2_cm)
print("B1 B2 =", b2_b1)
print("P1 B2 =", b2_p1)
# Get diff coeffs for b1 and p1
b1_Dg_fine = b1_ij / (b2_b1**0.5 * b1_flux)
p1_Dg_fine = p1_ij / (b2_p1**0.5 * p1_flux)
# Condense to two groups with new fluxes:
b1_Rabsxs_2gr = Condense2gr(rabsxs, b1_flux, energy, cutoffE=0.625)
b1_Diff_2gr = Condense2gr(b1_Dg_fine, b1_flux, energy, cutoffE=0.625)
p1_Rabsxs_2gr = Condense2gr(rabsxs, p1_flux, energy, cutoffE=0.625)
p1_Diff_2gr = Condense2gr(p1_Dg_fine, p1_flux, energy, cutoffE=0.625)
cm_Rabsxs_2gr = Condense2gr(rabsxs, cm_flx, energy, cutoffE=0.625)
cm_Diff_2gr = Condense2gr(1.0/3.0/cmmTransp, cm_flx, energy, cutoffE=0.625)
print()
print("B1 diff =", b1_Diff_2gr)
print("P1 diff =", p1_Diff_2gr)
print("CM diff =", cm_Diff_2gr)
print("B1 RABS =", b1_Rabsxs_2gr)
print("P1 RABS =", p1_Rabsxs_2gr)
print("CM RABS =", cm_Rabsxs_2gr)
SERPENT Serpent 2.2.1 found in ./serpent_HW3/fa2D_70gr_inf_res.m, but version 2.1.32 is defined in settings
Attempting to read anyway. Please report strange behaviors/failures to developers.
iterating 0
iterating 1
iterating 2
iterating 3
iterating 4
B2 = 0.00024591434041290686
CM B2 = 0.00024591434041290686
B1 B2 = 0.0002548806438124604
P1 B2 = 0.00025428519192265304
B1 diff = [1.69905265 0.88293977]
P1 diff = [1.70307681 0.88330774]
CM diff = [1.76303046 0.88447132]
B1 RABS = [0.00776182 0.06764274]
P1 RABS = [0.00776193 0.06764278]
CM RABS = [0.00776121 0.0676434 ]
# Plotting
plt.figure()
# Plot1d(energy, infinite_flx, xlabel="Energy, eV", ylabel='',
# fontsize=16, marker="-k", markerfill=False, markersize=6)
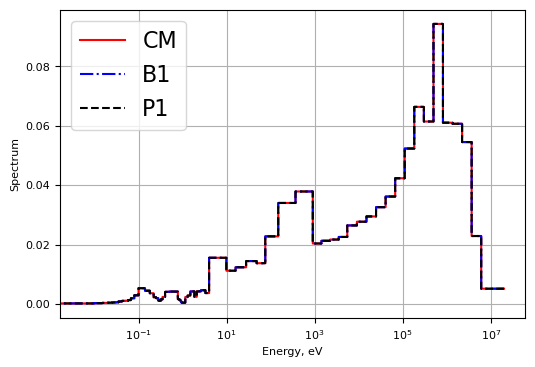
Plot1d(energy, cm_flx/cm_flx.sum(), xlabel="Energy, eV", ylabel='Spectrum',
fontsize=16, marker="-r", markerfill='none', markersize=6.5)
Plot1d(energy, b1_flux/b1_flux.sum(), xlabel="Energy, eV", ylabel='Spectrum',
fontsize=16, marker="-.b", markerfill='none', markersize=5)
Plot1d(energy, p1_flux/p1_flux.sum(), xlabel="Energy, eV", ylabel='Spectrum',
fontsize=16, marker="--k", markerfill='none', markersize=3)
plt.legend(['CM', "B1", "P1"])
<matplotlib.legend.Legend at 0x7f62ec0ca7b0>